Librerías informáticas utilizadas en análisis de imágenes dermatológicas con visión computacional: una revisión de literatura
DOI:
https://doi.org/10.51252/rcsi.v4i1.590Palabras clave:
clasificación dermatológica, diagnóstico clínico, inteligencia artificial, lesiones cutáneas, procesamiento de imágenes, segmentación de pielResumen
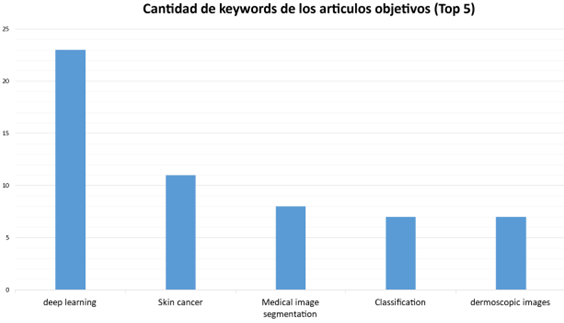
El análisis de imágenes cutáneas desempeña un papel fundamental en el ámbito de la dermatología, ya que posibilita la detección temprana y precisa de diversas afecciones de la piel. No obstante, este proceso se enfrenta a desafíos significativos debido a la variabilidad de características presentes en las lesiones cutáneas, tales como texturas, tonalidades y la existencia de vellosidades en el contorno. En este artículo, se presenta una revisión sistemática de literatura sobre librerías informáticas utilizadas en el análisis de imágenes dermatológicas con visión computacional. Esta investigación se basa en la declaración PRISMA y las bases de datos científicas: SCOPUS e IEEE Xplore para la búsqueda y tiene como objetivo identificar una amplia variedad de librerías informáticas y lesiones cutáneas. Los resultados mostraron 7 librerías y 21 lesiones dermatológicas, que contribuyen a un análisis más preciso y a un diagnóstico clínico más fiable para la detección oportuna de trastornos cutáneos. En conclusión, la presente investigación resalta librerías informáticas que tiene un impacto significativo en la mejora del diagnóstico clínico, lo cual es clave para el desarrollo de soluciones efectivas para la salud de las personas.
Citas
Abbas, Q., Ramzan, F., & Ghani, M. U. (2021). Acral melanoma detection using dermoscopic images and convolutional neural networks. Visual Computing for Industry, Biomedicine, and Art, 4(1). https://doi.org/10.1186/s42492-021-00091-z DOI: https://doi.org/10.1186/s42492-021-00091-z
Abdar, M., Samami, M., Dehghani Mahmoodabad, S., Doan, T., Mazoure, B., Hashemifesharaki, R., Liu, L., Khosravi, A., Acharya, U. R., Makarenkov, V., & Nahavandi, S. (2021). Uncertainty quantification in skin cancer classification using three-way decision-based Bayesian deep learning. Computers in Biology and Medicine, 135(April), 104418. https://doi.org/10.1016/j.compbiomed.2021.104418 DOI: https://doi.org/10.1016/j.compbiomed.2021.104418
Aladhadh, S., Alsanea, M., Aloraini, M., Khan, T., Habib, S., & Islam, M. (2022). An Effective Skin Cancer Classification Mechanism via Medical Vision Transformer. Sensors, 22(11). https://doi.org/10.3390/s22114008 DOI: https://doi.org/10.3390/s22114008
Albraikan, A. A., Nemri, N., Alkhonaini, M. A., Hilal, A. M., Yaseen, I., & Motwakel, A. (2023). Automated Deep Learning Based Melanoma Detection and Classification Using Biomedical Dermoscopic Images. Computers, Materials and Continua, 74(2), 2443–2459. https://doi.org/10.32604/cmc.2023.026379 DOI: https://doi.org/10.32604/cmc.2023.026379
Alcantud Marín, F., Alonso Esteban, Y., & Rico Bañón, D. (2015). Herramientas de cribado para la detección de retrasos o trastornos en el desarrollo: Una revisión sistemática de la literatura. Revista Española De Discapacidad, 3(2), 7–26. https://www.cedid.es/redis/index.php/redis/article/view/189 DOI: https://doi.org/10.5569/2340-5104.03.02.01
Alzubaidi, L., Fadhel, M. A., Al-Shamma, O., Zhang, J., Santamaría, J., & Duan, Y. (2022). Robust application of new deep learning tools: an experimental study in medical imaging. Multimedia Tools and Applications, 81(10), 13289–13317. https://doi.org/10.1007/s11042-021-10942-9 DOI: https://doi.org/10.1007/s11042-021-10942-9
Ashtari, P., Sima, D. M., De Lathauwer, L., Sappey-Marinier, D., Maes, F., & Van Huffel, S. (2023). Factorizer: A scalable interpretable approach to context modeling for medical image segmentation. Medical Image Analysis, 84(February 2022), 102706. https://doi.org/10.1016/j.media.2022.102706 DOI: https://doi.org/10.1016/j.media.2022.102706
Back, S., Lee, S., Shin, S., Yu, Y., Yuk, T., Jong, S., Ryu, S., & Lee, K. (2021). Robust Skin Disease Classification by Distilling Deep Neural Network Ensemble for the Mobile Diagnosis of Herpes Zoster. IEEE Access, 9, 20156–20169. https://doi.org/10.1109/ACCESS.2021.3054403 DOI: https://doi.org/10.1109/ACCESS.2021.3054403
Bala, D., Hossain, M. S., Hossain, M. A., Abdullah, M. I., Rahman, M. M., Manavalan, B., Gu, N., Islam, M. S., & Huang, Z. (2023). MonkeyNet: A robust deep convolutional neural network for monkeypox disease detection and classification. Neural Networks, 161, 757–775. https://doi.org/10.1016/j.neunet.2023.02.022 DOI: https://doi.org/10.1016/j.neunet.2023.02.022
Bibi, A., Khan, M. A., Javed, M. Y., Tariq, U., Kang, B. G., Nam, Y., Mostafa, R. R., & Sakr, R. H. (2022). Skin lesion segmentation and classification using conventional and deep learning based framework. Computers, Materials and Continua, 71(2), 2477–2495. https://doi.org/10.32604/cmc.2022.018917 DOI: https://doi.org/10.32604/cmc.2022.018917
Bing, S., Chawang, K., & Chiao, J.-C. (2023). A Tuned Microwave Resonant System for Subcutaneous Imaging. Sensors, 23(6), 3090. https://doi.org/10.3390/s23063090 DOI: https://doi.org/10.3390/s23063090
Caballé, N., Castillo, J. L., Gómez, J. A., Gómez, J. M., & Polo, M. (2020). Machine Learning Applied to Diagnosis of Human Diseases : A Systematic Review. Applied Sciences, 1–27. https://doi.org/10.3390/app10155135 DOI: https://doi.org/10.3390/app10155135
Cai, Y., Chen, H., Yang, X., Zhou, Y., & Cheng, K.-T. (2023). Dual-distribution discrepancy with self-supervised refinement for anomaly detection in medical images. Medical Image Analysis, 86, 102794. https://doi.org/10.1016/j.media.2023.102794 DOI: https://doi.org/10.1016/j.media.2023.102794
Cañedo, R., Rodriguez, R., & Marilis, M. (2010). Scopus : The largest database of peer-reviewed scientific literature available to underdeveloped countries Scopus : la mayor base de datos de literatura científica arbitrada al alcance de los países subdesarrollados Scopus : The largest database of peer-r. ACIMED, February 2016.
Cano, E., Mendoza-Avilés, J., Areiza, M., Guerra, N., Mendoza-Valdés, J. L., & Rovetto, C. A. (2021). Multi Skin Lesions Classification using Fine-tuning and Data-augmentation Applying Nasnet. PeerJ Computer Science, 7(Mcc), 1–20. https://doi.org/10.7717/PEERJ-CS.371 DOI: https://doi.org/10.7717/peerj-cs.371
Choudhary, P., Singhai, J., & Yadav, J. S. (2021). Curvelet and fast marching method-based technique for efficient artifact detection and removal in dermoscopic images. International Journal of Imaging Systems and Technology, 31(4), 2334–2345. https://doi.org/10.1002/ima.22633 DOI: https://doi.org/10.1002/ima.22633
Cui, R., Yang, R., Liu, F., & Geng, H. (2023). HD2A-Net: A novel dual gated attention network using comprehensive hybrid dilated convolutions for medical image segmentation. Computers in Biology and Medicine, 152(November 2022), 106384. https://doi.org/10.1016/j.compbiomed.2022.106384 DOI: https://doi.org/10.1016/j.compbiomed.2022.106384
Decharatanachart, P., Chaiteerakij, R., Tiyarattanachai, T., & Treeprasertsuk, S. (2021). Application of artificial intelligence in chronic liver diseases : a systematic review and meta ‑ analysis. BMC Gastroenterology, 1–16. https://doi.org/10.1186/s12876-020-01585-5 DOI: https://doi.org/10.1186/s12876-020-01585-5
Deng, Q., Beltran, J. C. C., & Lee, D. H. (2021). Assessment of Segmentation Impact on Melanoma Classification Using Convolutional Neural Networks. Journal of Computing Science and Engineering, 15(3), 115–124. https://doi.org/10.5626/JCSE.2021.15.3.115 DOI: https://doi.org/10.5626/JCSE.2021.15.3.115
Gálvez, A., Iglesias, A., Fister, I., Otero, C., & Díaz, J. A. (2021). NURBS functional network approach for automatic image segmentation of macroscopic medical images in melanoma detection. Journal of Computational Science, 56(April). https://doi.org/10.1016/j.jocs.2021.101481 DOI: https://doi.org/10.1016/j.jocs.2021.101481
Gu, R., Wang, G., Song, T., Huang, R., Aertsen, M., Deprest, J., Ourselin, S., Vercauteren, T., & Zhang, S. (2021). CA-Net: Comprehensive Attention Convolutional Neural Networks for Explainable Medical Image Segmentation. IEEE Transactions on Medical Imaging, 40(2), 699–711. https://doi.org/10.1109/TMI.2020.3035253 DOI: https://doi.org/10.1109/TMI.2020.3035253
Han, Z., Huang, H., Lu, D., Fan, Q., Ma, C., Chen, X., Gu, Q., & Chen, Q. (2023). One-stage and lightweight CNN detection approach with attention: Application to WBC detection of microscopic images. Computers in Biology and Medicine, 154(October 2022), 106606. https://doi.org/10.1016/j.compbiomed.2023.106606 DOI: https://doi.org/10.1016/j.compbiomed.2023.106606
He, S., Feng, Y., Grant, P. E., & Ou, Y. (2023). Segmentation ability map: Interpret deep features for medical image segmentation. Medical Image Analysis, 84(December 2022), 102726. https://doi.org/10.1016/j.media.2022.102726 DOI: https://doi.org/10.1016/j.media.2022.102726
Islam, M. M., Yang, H. C., Poly, T. N., Jian, W. S., & (Jack) Li, Y. C. (2020). Deep learning algorithms for detection of diabetic retinopathy in retinal fundus photographs: A systematic review and meta-analysis. Computer Methods and Programs in Biomedicine, 191, 1–16. https://doi.org/10.1016/j.cmpb.2020.105320 DOI: https://doi.org/10.1016/j.cmpb.2020.105320
Jaisakthi, S. M., Mirunalini, P., Aravindan, C., & Appavu, R. (2022). Classification of skin cancer from dermoscopic images using deep neural network architectures. Multimedia Tools and Applications, 15763–15778. https://doi.org/10.1007/s11042-022-13847-3 DOI: https://doi.org/10.1007/s11042-022-13847-3
Jiji, G. W., Rajesh, A., & Raj, P. J. D. (2021). CBI + R: A Fusion Approach to Assist Dermatological Diagnoses. International Journal of Image and Graphics, 21(1). https://doi.org/10.1142/S0219467821500054 DOI: https://doi.org/10.1142/S0219467821500054
Karri, M., Annavarapu, C. S. R., & Acharya, U. R. (2023). Skin lesion segmentation using two-phase cross-domain transfer learning framework. Computer Methods and Programs in Biomedicine, 231. https://doi.org/10.1016/j.cmpb.2023.107408 DOI: https://doi.org/10.1016/j.cmpb.2023.107408
Kosgiker, G. M., Deshpande, A., & Kauser, A. (2021). SegCaps: An efficient SegCaps network-based skin lesion segmentation in dermoscopic images. International Journal of Imaging Systems and Technology, 31(2), 874–894. https://doi.org/10.1002/ima.22545 DOI: https://doi.org/10.1002/ima.22545
Kumar, K. S., Suganthi, N., Muppidi, S., & Kumar, B. S. (2022). FSPBO-DQN: SeGAN based segmentation and Fractional Student Psychology Optimization enabled Deep Q Network for skin cancer detection in IoT applications. Artificial Intelligence in Medicine, 129(October 2021), 102299. https://doi.org/10.1016/j.artmed.2022.102299 DOI: https://doi.org/10.1016/j.artmed.2022.102299
La Salvia, M., Torti, E., Leon, R., Fabelo, H., Ortega, S., Martinez-Vega, B., Callico, G. M., & Leporati, F. (2022). Deep Convolutional Generative Adversarial Networks to Enhance Artificial Intelligence in Healthcare: A Skin Cancer Application. Sensors, 22(16). https://doi.org/10.3390/s22166145 DOI: https://doi.org/10.3390/s22166145
Lai, H., Fu, S., Zhang, J., Cao, J., Feng, Q., Lu, L., & Huang, M. (2022). Prior Knowledge-Aware Fusion Network for Prediction of Macrovascular Invasion in Hepatocellular Carcinoma. IEEE Transactions on Medical Imaging, 41(10), 2644–2657. https://doi.org/10.1109/TMI.2022.3167788 DOI: https://doi.org/10.1109/TMI.2022.3167788
Lan, Z., Cai, S., He, X., & Wen, X. (2022). FixCaps: An Improved Capsules Network for Diagnosis of Skin Cancer. IEEE Access, 10(May), 76261–76267. https://doi.org/10.1109/ACCESS.2022.3181225 DOI: https://doi.org/10.1109/ACCESS.2022.3181225
Lei, J., Yang, G., Wang, S., Feng, Z., & Liang, R. (2023). Category-aware feature attribution for Self-Optimizing medical image classification. Displays, 77(February), 102397. https://doi.org/10.1016/j.displa.2023.102397 DOI: https://doi.org/10.1016/j.displa.2023.102397
Li, S., Xie, Y., Wang, G., Zhang, L., & Zhou, W. (2022). Attention guided discriminative feature learning and adaptive fusion for grading hepatocellular carcinoma with Contrast-enhanced MR. Computerized Medical Imaging and Graphics, 97(February), 102050. https://doi.org/10.1016/j.compmedimag.2022.102050 DOI: https://doi.org/10.1016/j.compmedimag.2022.102050
Liberati, A., Altman, D. G., Tetzlaff, J., Mulrow, C., Gøtzsche, P. C., Ioannidis, J. P. A., Clarke, M., Devereaux, P. J., Kleijnen, J., & Moher, D. (2009). The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. In Journal of clinical epidemiology (Vol. 62, Issue 10). https://doi.org/10.1016/j.jclinepi.2009.06.006 DOI: https://doi.org/10.1016/j.jclinepi.2009.06.006
Liu, Z., Xiong, R., & Jiang, T. (2023). CI-Net: Clinical-Inspired Network for Automated Skin Lesion Recognition. IEEE Transactions on Medical Imaging, 42(3), 619–632. https://doi.org/10.1109/TMI.2022.3215547 DOI: https://doi.org/10.1109/TMI.2022.3215547
Lou, A., Guan, S., & Loew, M. (2023). CFPNet-M: A light-weight encoder-decoder based network for multimodal biomedical image real-time segmentation. Computers in Biology and Medicine, 154(December 2022), 106579. https://doi.org/10.1016/j.compbiomed.2023.106579 DOI: https://doi.org/10.1016/j.compbiomed.2023.106579
Mansour, R. F., Althubiti, S. A., & Alenezi, F. (2022). Computer Vision with Machine Learning Enabled Skin Lesion Classification Model. Computers, Materials and Continua, 73(1), 849–864. https://doi.org/10.32604/cmc.2022.029265 DOI: https://doi.org/10.32604/cmc.2022.029265
Maqsood, S., & Damaševičius, R. (2023). Multiclass skin lesion localization and classification using deep learning based features fusion and selection framework for smart healthcare. Neural Networks, 160, 238–258. https://doi.org/10.1016/j.neunet.2023.01.022 DOI: https://doi.org/10.1016/j.neunet.2023.01.022
Morgado, A. C., Andrade, C., Teixeira, L. F., & Vasconcelos, M. J. M. (2021). Incremental learning for dermatological imaging modality classification. Journal of Imaging, 7(9). https://doi.org/10.3390/jimaging7090180 DOI: https://doi.org/10.3390/jimaging7090180
Nawaz, M., Masood, M., Javed, A., Iqbal, J., Nazir, T., Mehmood, A., & Ashraf, R. (2021). Melanoma localization and classification through faster region-based convolutional neural network and SVM. Multimedia Tools and Applications, 80(19), 28953–28974. https://doi.org/10.1007/s11042-021-11120-7 DOI: https://doi.org/10.1007/s11042-021-11120-7
Nguyen, D. M. H., Nguyen, T. T., Vu, H., Pham, Q., Nguyen, M. D., Nguyen, B. T., & Sonntag, D. (2022). TATL: Task agnostic transfer learning for skin attributes detection. Medical Image Analysis, 78, 102359. https://doi.org/10.1016/j.media.2022.102359 DOI: https://doi.org/10.1016/j.media.2022.102359
Palacios, D., & Díaz, A. (2017). Dermatoscopia para principiantes ( i ): características generales. Medicina de Familia SEMERGEN, 43(3), 216–221. https://doi.org/10.1016/j.semerg.2015.11.009 DOI: https://doi.org/10.1016/j.semerg.2015.11.009
Pereira, P. M. M., Thomaz, L. A., Tavora, L. M. N., Assuncao, P. A. A., Fonseca-Pinto, R. M., Paiva, R. P., & Faria, S. M. M. d. (2022). Melanoma classification using light-Fields with morlet scattering transform and CNN: Surface depth as a valuable tool to increase detection rate. Medical Image Analysis, 75, 102254. https://doi.org/10.1016/j.media.2021.102254 DOI: https://doi.org/10.1016/j.media.2021.102254
Phan, D. T., Ta, Q. B., Ly, C. D., Nguyen, C. H., Park, S., Choi, J., Se, H. O., & Oh, J. (2023). Smart Low Level Laser Therapy System for Automatic Facial Dermatological Disorder Diagnosis. IEEE Journal of Biomedical and Health Informatics, 27(3), 1546–1557. https://doi.org/10.1109/jbhi.2023.3237875 DOI: https://doi.org/10.1109/JBHI.2023.3237875
Qiu, S., Li, C., Feng, Y., Zuo, S., Liang, H., & Xu, A. (2023). GFANet: Gated Fusion Attention Network for skin lesion segmentation. Computers in Biology and Medicine, 155(December 2022). https://doi.org/10.1016/j.compbiomed.2022.106462 DOI: https://doi.org/10.1016/j.compbiomed.2022.106462
Quero-Caiza, W., & Altuve, M. (2021). Recognition of Skin Lesions in Dermoscopic Images using Local Binary Patterns and Multinomial Logistic Regression. IEEE Latin America Transactions, 20(7), 2020–2028. https://doi.org/10.1109/TLA.2021.9827475 DOI: https://doi.org/10.1109/TLA.2021.9827475
Szolga, L., Bozga, D., & Florea, C. (2001). End-User Skin Analysis (Moles) through Image Acquisition and Processing System. Sensors, 20, 9–11.
Thurnhofer-Hemsi, K., Lopez-Rubio, E., Dominguez, E., & Elizondo, D. A. (2021). Skin lesion classification by ensembles of deep convolutional networks and regularly spaced shifting. IEEE Access, 9, 112193–112205. https://doi.org/10.1109/ACCESS.2021.3103410 DOI: https://doi.org/10.1109/ACCESS.2021.3103410
Wang, K., Zhang, X., Lu, Y., Zhang, W., Huang, S., & Yang, D. (2023). GSAL: Geometric structure adversarial learning for robust medical image segmentation. Pattern Recognition, 140, 109596. https://doi.org/10.1016/j.patcog.2023.109596 DOI: https://doi.org/10.1016/j.patcog.2023.109596
Wei zhu, Liu, L., Kuang, F., Li, L., Xu, S., & Liang, Y. (2022). An efficient multi-threshold image segmentation for skin cancer using boosting whale optimizer. Computers in Biology and Medicine, 151(PA), 106227. https://doi.org/10.1016/j.compbiomed.2022.106227 DOI: https://doi.org/10.1016/j.compbiomed.2022.106227
Wu, H., Pan, J., Li, Z., Wen, Z., & Qin, J. (2021). Automated Skin Lesion Segmentation Via an Adaptive Dual Attention Module. IEEE Transactions on Medical Imaging, 40(1), 357–370. https://doi.org/10.1109/TMI.2020.3027341 DOI: https://doi.org/10.1109/TMI.2020.3027341
Wu, Yang, H., Peng, L., Lian, Z., Li, M., Qu, G., Jiang, S., & Han, Y. (2022). AGNet: Automatic generation network for skin imaging reports. Computers in Biology and Medicine, 141(June 2021), 105037. https://doi.org/10.1016/j.compbiomed.2021.105037 DOI: https://doi.org/10.1016/j.compbiomed.2021.105037
Yang, C., & Lu, G. M. (2022). Skin Lesion Segmentation with Codec Structure Based Upper and Lower Layer Feature Fusion Mechanism. KSII Transactions on Internet and Information Systems, 16(1), 60–79. https://doi.org/10.3837/tiis.2022.01.004 DOI: https://doi.org/10.3837/tiis.2022.01.004
Yao, P., Shen, S., Xu, M., Liu, P., Zhang, F., Xing, J., Shao, P., Kaffenberger, B., & Xu, R. X. (2022). Single Model Deep Learning on Imbalanced Small Datasets for Skin Lesion Classification. IEEE Transactions on Medical Imaging, 41(5), 1242–1254. https://doi.org/10.1109/TMI.2021.3136682 DOI: https://doi.org/10.1109/TMI.2021.3136682
Yilmaz, E., & Trocan, M. (2021). A modified version of GoogLeNet for melanoma diagnosis. Journal of Information and Telecommunication, 5(3), 395–405. https://doi.org/10.1080/24751839.2021.1893495 DOI: https://doi.org/10.1080/24751839.2021.1893495
Zaid, M., Ali, S., Ali, M., Hussein, S., Saadia, A., & Sultani, W. (2022). Identifying out of distribution samples for skin cancer and malaria images. Biomedical Signal Processing and Control, 78(May), 103882. https://doi.org/10.1016/j.bspc.2022.103882 DOI: https://doi.org/10.1016/j.bspc.2022.103882
Zhang, J., Liu, Y., Wu, Q., Wang, Y., Liu, Y., Xu, X., & Song, B. (2022). SWTRU: Star-shaped Window Transformer Reinforced U-Net for medical image segmentation. Computers in Biology and Medicine, 150(August), 105954. https://doi.org/10.1016/j.compbiomed.2022.105954 DOI: https://doi.org/10.1016/j.compbiomed.2022.105954
Zhao, D., Qi, A., Yu, F., Heidari, A. A., Chen, H., & Li, Y. (2023). Multi-strategy ant colony optimization for multi-level image segmentation: Case study of melanoma. Biomedical Signal Processing and Control, 83(February), 104647. https://doi.org/10.1016/j.bspc.2023.104647 DOI: https://doi.org/10.1016/j.bspc.2023.104647
Zhao, H., Wang, A., & Zhang, C. (2022). Research on melanoma image segmentation by incorporating medical prior knowledge. PeerJ Computer Science, 8. https://doi.org/10.7717/PEERJ-CS.1122 DOI: https://doi.org/10.7717/peerj-cs.1122
Publicado
Cómo citar
Número
Sección
Licencia
Derechos de autor 2024 Jose Carlos Huanatico-Lipa, Marco Antonio Coral-Ygnacio

Esta obra está bajo una licencia internacional Creative Commons Atribución 4.0.
Los autores retienen sus derechos:
a. Los autores retienen sus derechos de marca y patente, y tambien sobre cualquier proceso o procedimiento descrito en el artículo.
b. Los autores retienen el derecho de compartir, copiar, distribuir, ejecutar y comunicar públicamente el articulo publicado en la Revista Científica de Sistemas e Informática (RCSI) (por ejemplo, colocarlo en un repositorio institucional o publicarlo en un libro), con un reconocimiento de su publicación inicial en la RCSI.
c. Los autores retienen el derecho a hacer una posterior publicación de su trabajo, de utilizar el artículo o cualquier parte de aquel (por ejemplo: una compilación de sus trabajos, notas para conferencias, tesis, o para un libro), siempre que indiquen la fuente de publicación (autores del trabajo, revista, volumen, número y fecha).